Functional analysis
The development of high-throughput measurements brought an unprecedented increase in the number of biomedical variables. The decreasing cost of measurement and computation in the data analysis, together with the hypothesis free study design, make the interpretation phase more and more crucial, in many projects the interpretation phase practically became the bottleneck. As a response to the very large numbers of variables many data analysis methods have appeared to focus on set of variables. These sets are defined by the codified general or even by the domain-specific background knowledge, for example based on gene annotations, taxonomies, ontologies, gene mappings between organisms, pathway knowledge bases.
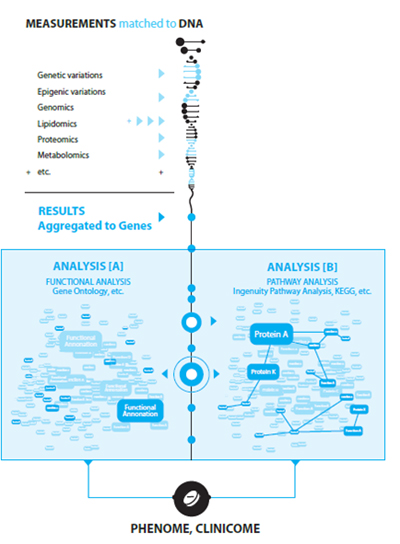
We offer a unique solution for the interpretational bottleneck, which incorporates wide range of background knowledge and utilities as well:
-
The entities of the data analysis can be embedded in a probabilistic logical knowledge base. This knowledge base contains the following interrelated information sources:
- Free-text, hyperlinked disease specific knowledge bases,
- Automatically extracted and manually curated knowledge bases using controlled language and vocabulary,
- Logical knowledge bases,
- Dependency maps and causal models.
- The conclusions are interpreted as (reporting) actions within a decision theoretic framework, for which utilities can be specified as well.


